
Georg Hausner
Fungi and Mitochondria
Zubaer, A., Wai, A., and *Hausner, G. 2025. Comparative mitogenomics of Leptographium procerum, Leptographium terebrantis and Leptographium wingfieldii, an invasive fungal species in Canadian forests. Can. J. Micro. http://dx.doi.org/10.1139/cjm-2024-0179
Bhunjun, C.S. et al. (Jankowiak R. and Hausner G.)* 2024. What are the 100 most cited fungal genera? Studies in Mycology. (contribution section 96: Ophiostoma; pages 259-266)*. Studies in Mycology 108: (In press).
Mukhopadhyay, J., and Hausner, G. 2024. Interconnected roles of fungal nuclear and intron encoded maturases: At the crossroads of mitochondrial intron splicing. Biochemistry and Cell Biology. http://dx.doi.org/10.1139/bcb-2024-0046 (accepted May/2024).
Kulik T., Van Diepeningen A.D., *Hausner G. 2023. Editorial: The Significance of Mitogenomics in Mycology, Volume II. Frontiers in Microbiology. 14, 1344877. doi: 10.3389/fmicb.2023.1344877
103. Mukhopadhyay, J., Wai, A., *Hausner, G. 2023. The mitogenomes of Leptographium aureum, Leptographium sp., and Grosmannia fruticeta: expansion by introns. Frontiers in Microbiology. 14, 1240407. https://doi.org/10.3389/fmicb.2023.1240407
Wai A, *Hausner G. 2022. The compact mitochondrial genome of Ceratocystiopsis pallidobrunnea. Can. J. Microbiol. 68(9):569-575. https://doi.org/10.1139/cjm-2022-0038
Jankowiak R, Solheim H*, Bilański P, Mukhopadhyay J, Hausner G. 2022. Ceratocystiopsis spp. associated with pine- and spruce-infesting bark beetles in Norway. Mycological Progress 21: 61. https://doi.org/10.1007/s11557-022-01808-x
Mukhopadhyay J, Wai, A., Hutchison LJ, *Hausner G. 2022. The mitogenome of Urnula criterium. Can. J. Microbiol. 68(8):561-568. https://doi.org/10.1139/cjm-2022-0012
Kouvelis VN, Hausner G. 2022. Editorial: Mitochondrial genomes and mitochondrion related genes provide insights to fungal evolution. Frontiers in Microbiology. 13:897981. doi: 10.3389/fmicb.2022.897981 Part of an ebook: Kouvelis, V. N., Hausner, G., eds. (2022). Mitochondrial Genomes and Mitochondrion Related Gene Insights to Fungal Evolution. Lausanne: Frontiers Media SA. doi: 10.3389/978-2-88976-061-9 (10 articles).
Mukhopadhyay J, *Hausner G. 2021. Organellar introns in fungi, algae and plants. Cells. 10(8): 2001. doi: 10.3390/cells10082001.
Zubaer A, Wai A, Patel N, Perillo J and Hausner G (2021) The Mitogenomes of Ophiostoma minus and Ophiostoma piliferum and Comparisons With Other Members of the Ophiostomatales. Front. Microbiol. 12:618649. doi: 10.3389/fmicb.2021.618649
Thank you to our collaborators for allowing us to participate in their exciting work:
Mayers CG, Harrington TC, Wai A and Hausner G (2021) Recent and Ongoing Horizontal Transfer of Mitochondrial Introns Between Two Fungal Tree Pathogens. Front. Microbiol. 12:656609. doi: 10.3389/fmicb.2021.656609
Alvan Wai, Chen Shen, Carta Andrell, Alexandra Dansen, Pedro W. Crous, Georg Hausner. 2019. Intron-encoded ribosomal proteins and N-acetyltransferases within the mitogenomes of some fungi: Here today, gone tomorrow? Mitochondrial DNA part A. https://www.tandfonline.com/eprint/xH78GN9rkfjtSEZKCyNG/full?target=10.1080/24701394.2019.1580272


The mitochondrial rps3 gene is usually encoded by an rnl group I intron in many filamentous ascomycetes fungi. However, there are groups where this gene is free standing or has relocated to the nuclear genome (some members of the Capnodiales).
Mitochondrial
clade of NAT
N-acetyltransferase (NAT) phylogeny and Ascomycota rps3 location
A copy of NAT appears to have transferred in some fungi from the nuclear genome to the mitochondrial genome.
In some fungi a copy of the rps3 gene appears to have transferred from the mitochondrial genome to the nuclear genome.
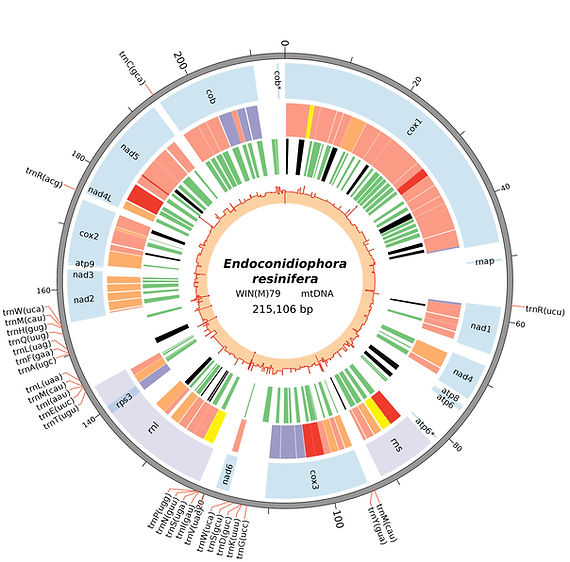
Zubaer A, Wai A, Hausner G. 2018. The mitochondrial genome of Endoconidiophora resinifera is intron rich. Sci Rep. 8(1):17591. doi:10.1038/s41598-018-35926-y.
The fate of composite elements such as introns plus IEPs (I+H+). The composite element invades an empty site and from here it could spread into other sites (only site B shown for simplicity) and related IEPs could still interact with their ancestral intron version possibly facilitating splicing or mobility thus enhancing the chances of the ancestral intron to persist. This form of mutualism could even complement situations where the ancestral intron ORF has started to accumulate deleterious mutations (H-). Other composite elements may be strictly subject to drift and first the ORF is degenerating and eventually the intron is lost from the genome and possibly from the population. There might be situations where the composite elements have been co-opted as maturases or regulatory elements or as platforms for expressing essential genes (rps3) and these introns would be subject to adaptive selection and thus could become fixed in the genome and the population (Figure adapted from Gogarten and Hilario [83]).

Engelberg, R., Danielson, A., Wang, S., Singh, M., Wai, A., Duan, K., Sorensen, J., Hausner, G., and Kumar, A. 2018. Creation of a drug-sensitive reporter strain of Pseudomonas aeruginosa as tool for the rapid screening of antimicrobial products. Journal of Microbiological Methods.
Collaborative work: Ayush Kumar and Georg Hausner.

Abboud TG, Zubaer A, Wai A, Hausner G. 2018. The complete mitochondrial genome of the Dutch elm disease fungus Ophiostoma novo-ulmi subspecies novo-ulmi. Can. J. Micro. http://www.nrcresearchpress.com/eprint/5GHkkrFn5tNtg8s3RFVp/full

Guha*, T.K., Wai*, A., Mullineux, S.-T., and Hausner, G. 2017. The intron landscape of the mtDNA cytb gene among the Ascomycota: introns and intron-encoded open reading frames. Mitochondrial DNA part A. http://dx.doi.org/10.1080/24701394.2017.1404042. [*equal contribution].


Bilto IM, Guha TK, Wai A, Hausner G. 2017. Three new active members of the I-OnuI family of homing endonucleases. Can. J. Micro. Doi: 10.1139/cjm-2017-0067.


Guha, T.K., Wai, A., Hausner, G. 2017. Programmable genome editing tools and their regulation for efficient genome engineering. Computational and Structural Biotechnology Journal. 15: 146-160, doi: 10.1016/j.csbj.2016.12.006.

Examples of programmable genome editing tools. (a) Single-motif LAGLIDADG homing endonucleases, (b) double-motif LAGLIDADG homing endonucleases, (c) megaTAL, (d) MegaTev, (e) zinc-finger nucleases (ZFN), (f) transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated proteins (Cas) systems using (g) Cas9 or (h) Cpf1, (i) targetrons, (j) triplex-forming oligonucleotide (TFO) nucleases, and (k) structure-guided nucleases (SGNs).
(Hafez and Hausner 2012; Guha et al. 2017)
Hafez, M., Hausner, G., 2017. Intron Homing. IN: Elsevier Reference Module in Life Sciences. ISBN: 978-0-12-809633-8, http://dx.doi.org/10.101/B978-0-12-809633-8.06604-8.
Guha, T.K., Hausner, G. 2017. Insertion of ribozyme based switches into homing endonuclease genes. Methods in Molecular Biology - In Vitro Mutagenesis: Methods and Protocols; ed. Reeves A. Invited Book Chapter. Springer Verlag. Volume 1488: 135-152.
Bilto, I.M., Hausner, G., 2016. The diversity of mtDNA rns introns among strains of Ophiostoma piliferum, Ophiostoma pluriannulatum and related species. SpringerPlus 5:1408
Guha, T.K., Hausner, G. 2016. Using group II introns for attenuating the in vitro and in vivo expression of a homing endonuclease. PLOS ONE (Feb. 24th/2016) doi:10.1371/journal.pone.0150097

In Chaetomium thermophilum (DSM 1495) within the mitochondrial DNA (mtDNA) small ribosomal subunit (rns) gene a group IIA1 intron interrupts an open reading frame (ORF) encoded within a group I intron (mS1247). This arrangement offers the opportunity to examine if the nested group II intron could be utilized as a regulatory element for the expression of the homing endonuclease (HEase). Constructs were generated where the codon-optimized ORF was interrupted with either the native group IIA1 intron or a group IIB type intron. This study showed that the expression of the HEase (in vivo) in Escherichia coli can be regulated by manipulating the splicing efficiency of the HEase ORF-embedded group II introns. Exogenous magnesium chloride (MgCl2) stimulated the expression of a functional HEase but the addition of cobalt chloride (CoCl2) to growth media antagonized the expression of HEase activity. Ultimately the ability to attenuate HEase activity might be useful in precision genome engineering, minimizing off target activities, or where pathways have to be altered during a specific growth phase.
Hafez, M., and Hausner, G. 2015. Convergent evolution of twintron-like configurations: One is never enough. RNA Biol. 12(12): 1275-1288.
http://www.tandfonline.com/doi/pdf/10.1080/15476286.2015.1103427

Introns inserted within introns are commonly referred to as twintrons, however the original definition for twintron implied that splicing of the external member of the twintron could only proceed upon splicing of the internal member. This review examines the various types of twintron-like arrangements that have been reported and assigns them to either nested or twintron categories that are subdivided further into subtypes based on differences of their mode of splicing. Twintron-like arrangements evolved independently by fortuitous events among different types of introns but once formed they offer opportunities for the evolution of new regulatory strategies and/or novel genetic elements.
Guha, T.K., and Hausner G. 2014. A homing endonuclease with a switch: Characterization of a twintron encoded homing endonuclease. Fungal Genetics and Biology, 65: 57-68.
http://www.sciencedirect.com/science/article/pii/S1087184514000061

The small ribosomal subunit gene residing in the mitochondrial DNA of the thermophilic fungus Chaetomium thermophilum var. thermophilum La Touche DSM 1495 is interrupted by a twintron at position mS1247. The mS1247 twintron represents the first mixed twintron found in fungal mtDNA, composed of an external group I intron encoding a LAGLIDADG open reading frame that is interrupted by an internal group II intron. Splicing of the internal group II intron reconstitutes the open reading frame and thus facilitates the expression of the encoded homing endonuclease. The cleavage assays suggest that the twintron encodes an active homing endonuclease that could potentially mobilize the twintron to rns genes that have not yet been invaded by this mobile composite element.
Hafez, M., Majer, A., Sethuraman, J., Rudski, S.M., Michel F., and Hausner, G. 2013. The mtDNA rns gene landscape in the Ophiostomatales and other fungal taxa: twintrons, introns, and intron-encoded proteins. Fungal Genetics and Biology. 53: 71-83.

Comparative sequence analysis of the mitochondrial small subunit ribosomal RNA (rns) gene among species of Ophiostoma, Grosmannia, Ceratocystiopsis and related taxa provides an overview of the types of introns that have invaded this gene within the ophiostomatoid fungi. The rns gene appears to be a reservoir for a number of group I and group II introns along with intron-associated open reading frames such as homing endonucleases and reverse transcriptases. This study uncovered two twintrons, one at position mS917 where a group ID intron encoding LAGLIDADG ORF invaded another ORF-less group ID intron. Another twintron complex was detected at position mS1247 here a group IIA1 intron invaded the open reading frame embedded within a group IC2 intron. Overall the distribution of the introns does not appear to follow evolutionary lineages suggesting the possibility of rare horizontal gains and frequent losses. Results of this study will make a significant contribution to the understanding of the complexity of the mitochondrial intron landscape, and offer a resource to those annotating mitochondrial genomes. It will also serve as a resource to those that bioprospect for ribozymes and homing endonucleases.
Hausner G., Hafez, M., Edgell, D.R. 2014. Bacterial group I introns: mobile RNA catalysts. Mobile DNA, 5:8
Sethuraman, J., Rudski, S.M., Wosnitza, K.M, Hafez, M., Guppy, B., *Hausner, G. 2013. Evolutionary dynamics of introns and their open reading frames in the U7 region of the mitochondrial rnl gene in species of Ceratocystis Fungal Biology, 117: 791-806.
Hafez, M., and *Hausner G. 2012. Homing endonucleases: DNA scissors on a mission. Genome 55(8): 553-569.
Hafez, M., and *Hausner G. 2011. Variable mitochondrial rns gene arrangement within strains of Ophiostoma minus. Fungal Biology 115: 1122-1137.
Hafez, M., *Hausner, G. 2011. Characterization of the O.ul-mS952 intron: a potential molecular marker to distinguish between Ophiostoma ulmi and Ophiostoma novo-ulmi subsp. americana. https://www.waset.org/author/georg-hausner World Academy of Science, Engineering and Technology Journal 59: 1767-1775.
Mullineux S-T., Costa M., Bassi, G.S., Michel, F., *Hausner, G. 2010. A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions. RNA 16:1818-1831.
Sethuraman J., Majer, A., Friedrich, N.C., Edgell, D.R., and *Hausner G. 2009. Genes-within-genes: Multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within a rnl group I intron of Ophiostoma and related taxa. Molecular Biology and Evolution. 26: 2299-2315.
Mullineux, S.-T. and *Hausner G. 2009. Evolution of rDNA ITS1 and ITS2 sequences and RNA secondary structures within members of the fungal genera Grosmannia and Leptographium. Fungal Genetics and Biology 46: 855-867.